Hi, I am Joshua Riback, a student in the Biophysical Sciences graduate program co-advised in both the Drummond and Sosnick Labs.
Recently we have published a paper characterizing an intersection between polymer (bio)physics and cell biology. Polymer biophysics elucidates the principles governing the organized demixing of molecules inside of cells. Protein and RNA can demix into massive assemblies such as gels, liquids, and fibers. They lack a fixed stoichiometry so we refer to them as quinary assemblies. We focus on the properties of assemblies composed of poly(A)-binding protein (Pab1) that form in eukaryotic cells in response to cellular stress. We find that the demixing of Pab1 has been tuned for rapid response to stress and is beneficial for surviving that stress. For a little more background, see Allan’s post on our paper.
For those who wish to dive into the paper, please proceed here. Since our story differs significantly from the current results in the field, I have focused this blog post on the polymer biophysics I expected would govern Pab1 demixing vs. reality.
Polymer biophysics of Pab1: Expectations
Prior to obtaining data and based on my reading of the literature in the field, the principles I expected were going to govern Pab1 demixing were:
-
Pab1 would have an intrinsically disordered region (‘IDR’ or non-foldable low complexity regions) that would allow different Pab1 molecules to flexibly interact in the quinary assembly.
-
Demixing would be favorable due to multivalent interactions between RNA and Pab1 and between IDRs on different Pab1 molecules.
-
Pab1’s IDR would promote demixing through interactions between charge, aromatic, or polar residues (i.e. coulomb interactions or hydrogen bonds) and not from aliphatic residues because they are depleted in IDRs. The interacting residues would be enriched in the composition of the IDR.
-
Pab1’s IDR would be collapsed. When demixed, Pab1’s IDR would interact with other Pab1 molecules; but when monomeric, Pab1’s IDR would self-interact causing it to collapse.
Polymer biophysics of Pab1: Reality
Expectation: #1
Pab1 would have an IDR that would allow different Pab1 molecules to flexibly interact in the quinary assembly.
Expectation: #2
Demixing would require multivalent interactions between RNA and Pab1 and/or between IDRs on different Pab1 molecules.
Expectation: #3
Pab1’s IDR would promote demixing through interactions between charge, aromatic, or polar residues (ie coulomb interactions or hydrogen bonds) and not from aliphatic residues because they are depleted in IDRs. The interacting residues would be enriched in the composition of the IDR.
IDRs are understood by their compositional properties. Based on known IDRs that can demix, important compositional features have been identified (see the review). One of these IDRs, the RGG domain is necessary for demixing of the P-granule protein, LAF-1. The composition of the P domain is different from that of the RGG domain and these other known IDRs implicated in demixing.
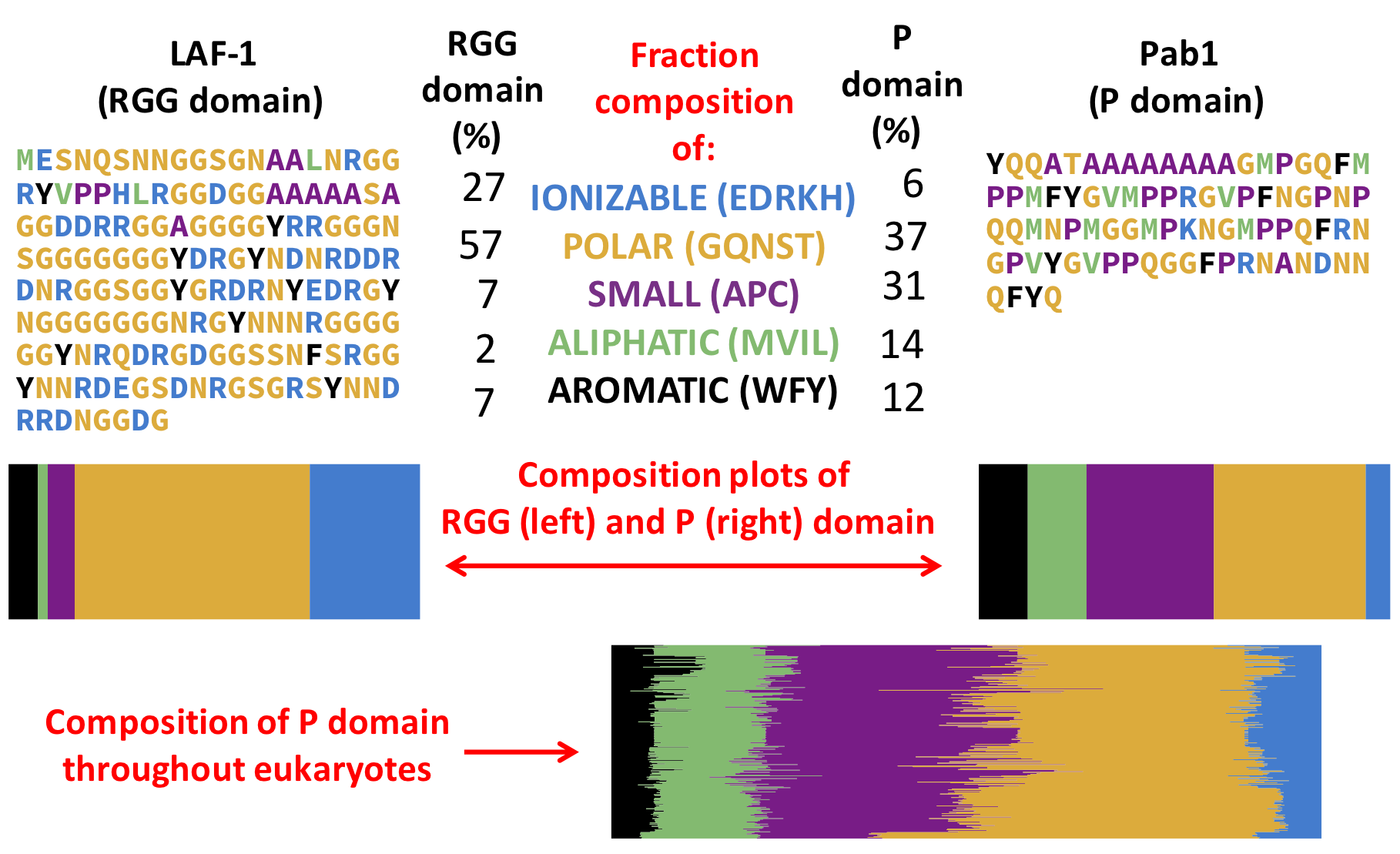
For clarity, the compositional plots are fraction residues of each type, and for the evolutionary composition plot, each row is a different species. The hydrophobicity of the residue groups are roughly aromatic, aliphatic, small, polar, and ionizable from most to least hydrophobic.
Expectation: #4
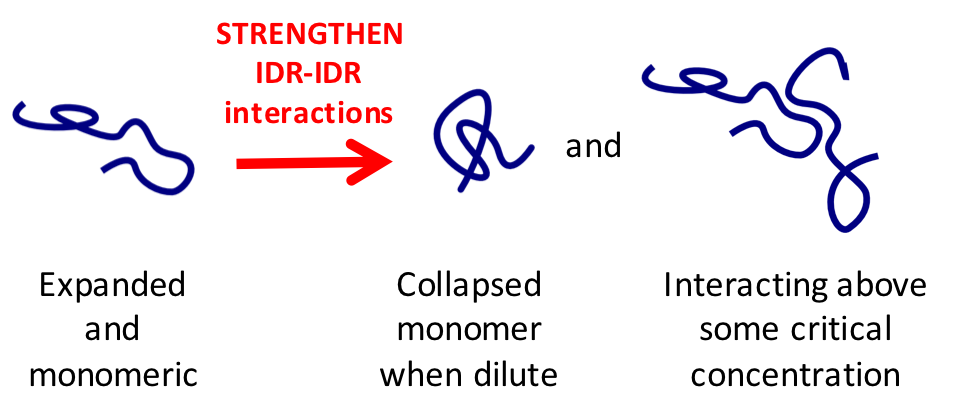
Pab1’s IDR would be collapsed. When demixed, Pab1’s IDR would interact with other Pab1 molecules, but when monomeric, Pab1’s IDR would self-interact causing it to collapse.
This is a concept drawn from polymer physics 101. If IDR-IDR interactions are sufficiently strong, then the IDR will either collapse if at low concentration or demix if at sufficiently high concentration.
Expectation vs. Reality Summary
| # |
Expectation |
Reality |
Expectation~Reality |
| 1 |
Pab1 has IDR |
IDR (P domain) is conserved in eukaryotes |
Yes |
| 2 |
Demixing requires RNA:Pab1 interactions and IDR:IDR interactions |
Demixing requires only folded domains |
No |
| 3 |
IDR promotes demixing with charge, aromatic, or polar residues |
Net hydrophobicity of amino acid residues enhances demixing |
No |
| 4 |
IDR is collapsed. Extent of IDR collapse would correlate with enhancing demixing |
Size of IDR correlates with the temperature of demixing in vivo and in vitro |
Yes |
Feel free to contact me with questions or comments.